Eimeria: The chicken parasite costing farmers billions
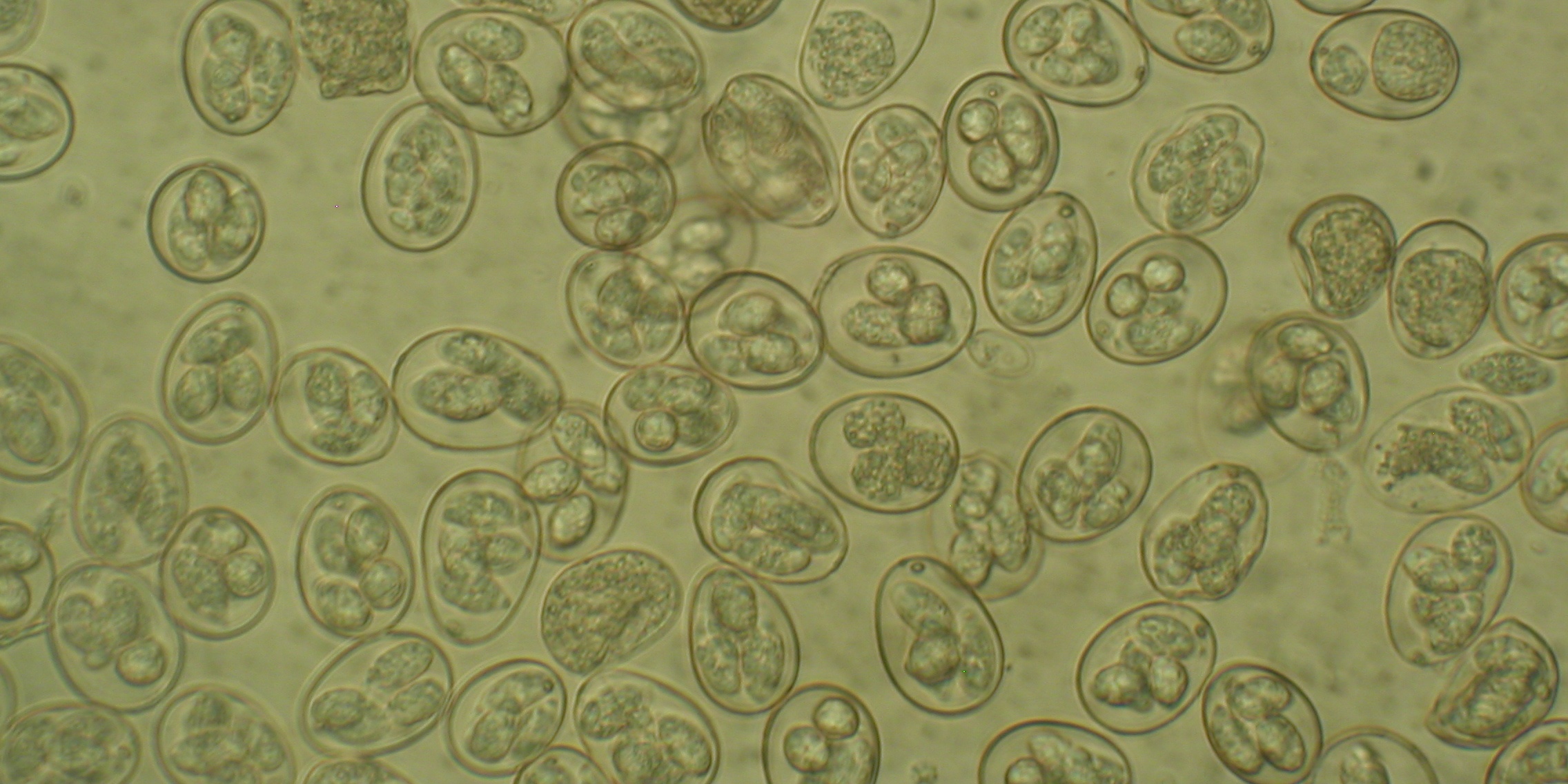
A high-quality genome of the coccidian parasite Eimeria tenella has been sequenced and published by scientists at the Sanger Institute. This single-celled organism causes an intestinal disease in chickens — called coccidiosis — which can lead to severe diarrhoea and death. This is both bad for the birds’ welfare and costs the global poultry industry billions every year.
Why are people interested in Eimeria genomes?
Although it poses no health risks to humans, E. tenella and six other species of Eimeria are very costly to the chicken farming industry. In 2016 in the UK alone, the cost of prevention, treatment and the losses from coccidiosis have been estimated at almost £100 million. Globally, that figure was somewhere between £7 and £13 billion, the equivalent of 16p per chicken produced.
There are drugs available, but the parasite becomes resistant to these fairly quickly. Live parasite vaccines can also be used, but cannot be produced at a scale or cost suitable for all chickens. It is hoped that understanding the genome will help produce a more comprehensive vaccine that protects against all the major species of Eimeria, and across different sectors of the poultry industry.

What do scientists hope to discover?
The seven species of Eimeria that infect chickens all target distinct zones of the gut and cause different types of disease. Curiously, Eimeria genetic diversity has been found to vary enormously between Eimeria populations. Currently we don’t know why this happens or how it impacts on control of disease. Intriguingly, three new cryptic Eimeria species have recently been discovered, showing a need for better understanding. Do each of the species have certain characteristics which help them fill different niches in the chicken gut? How are they evolving and can we use this information to keep ahead of this evolution?
Prof. Damer Blake at the Royal Veterinary College, collaborator on the Darwin Tree of Life project, has been investigating this phenomenon. The new genome sequence will aid his research. Consider the genome as a “parts list” for an organism, which lets us identify the genes that are worth investigating. Sometimes this can be a relatively small group of genes within the genome.
In this case, it helps explore differences between the types of Eimeria and their distinct characteristics — including their virulence, drug resistance and ability to escape from vaccine-induced immunity. Ultimately, this may help develop new vaccines.
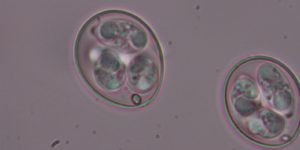
Why does this genome excite genomics experts?
Throughout Eimeria genomes we see very long sequences repeating the same amino acids. Although this also happens in proteins from other species, it is nowhere near the same extent.
Repeats like this should be very disruptive, changing the structure of proteins. We would therefore expect this to happen in certain types of genes, for example those that have only appeared recently in the organism’s evolutionary history or in genes that are less important to the organism’s functions. But in Eimeria they turn up everywhere, including some very ancient and critical proteins.
This phenomenon is only seen at this scale in Eimeria, and genomicists are keen to find out why.
What has studying this genome already taught us?
Protozoan parasites like Eimeria have all evolved to do the same basic things: invade the cells of their host, and survive attempts by the host’s immune system to destroy them. It’s an evolutionary race between host and parasite, sometimes described as the “Red Queen” hypothesis, based on a quote from Alice in Wonderland:

Earlier studies on shorter sequences of the Eimeria genomes showed that the most rapidly evolving genes, those involved most directly with the arms race between host and parasite, have a very different origin from the same sorts of genes in related species. This evolutionary battle has regularly resulted in parasites switching their weapons for ones which might give them the upper hand.
Scientists hope that by studying the genomes of more and more parasites, they can better understand how this switching takes place and why particular types of genes are useful for foiling the host immune system.
The challenges of sequencing single-celled organisms
This is the first protist (single-celled organism with a nucleus) to have its genome published in connection with the Darwin Tree of Life project. Eventually we will sequence thousands of protists, and several of our partners and other collaborators are currently focusing on this, including CCAP, the Earlham Institute, the Marine Biological Association and the University of Oxford.
However, these tiny organisms can be tricky to sequence. A single cell does not provide enough DNA for a high-quality genome sequence. While many species can be grown in the lab to large numbers, most must currently be caught in the wild, making it difficult to get large amounts of DNA. Part of this project’s ground-breaking work will be pioneering the technological advancements required to overcome these challenges, and making these openly available to the scientists of the future.
This article was based on an interview with Dr. Adam Reid, Senior Scientist at the Wellcome Sanger Institute, who has been researching the genomics of Eimeria and other parasites for the past decade.
The article was originally published on the Wellcome Sanger Institute blog.