New Tree of Life Gateway launches
When Carl Linnaeus began to formalise the cataloguing of all life on earth over 250 years ago, he developed a ranked taxonomy to organise our understanding of all species on earth. As time has passed and knowledge of diversity, biology and evolutionary theory has increased, the Linnaean classification system has evolved alongside it. This Linnaean catalogue is one of the crowning achievements of science: an openly accessible, globally treasured and communally owned record of all of the books in the great library of life. The next step in comprehending the interconnectedness of all life is to read and understand the material present in all these Linnaean books: decoding the genomes of all living organisms.
In 1977, Frederick Sanger and colleagues decoded the complete DNA sequence of a genome for the first time. The ΦX174 bacteriophage – a virus that infects E. coli – is made up of just 5386 DNA base pairs. This ground-breaking work would lay the foundations for dramatic advances in genomics, with the human genome – 3.1 billion DNA base pairs deciphered over 13 years – completed in 2003. Forty-four years since Sanger revealed the first genome sequence, the Tree of Life programme at the Wellcome Sanger Institute, the research institute that bears his name, aims to advance the field of genomics yet further. In our programme, the genome sequences of thousands of diverse species will be rapidly determined to the highest quality to generate references of unparalleled accuracy.
The Tree of Life programme uses DNA sequencing to examine the diversity of complex organisms all around us, assembling high-quality genome sequences that can be used to understand the evolution of life and to aid the conservation of biodiversity. The programme delivers its goals through major projects, especially the Darwin Tree of Life project. The Darwin project is a partnership with biodiversity and analysis colleagues throughout the UK, and aims to decipher the genome sequences of all approximately 70,000 eukaryotic organisms (plants, animals, fung and single-celled species) that live on and around Britain and Ireland.
We are also generating genomes for the Aquatic Symbiosis Genomics Project, which will sequence the 1000 participants of 500 symbioses between eukaryotic hosts and their symbiotic microbial ‘cobionts’ to understand their interconnected biology, reference vertebrate genomes as part of the Vertebrate Genomes Project, and other projects. Tree of Life is part of the Earth BioGenome Project, which has the modest goal of attempting to catalogue and sequence all complex life on earth.
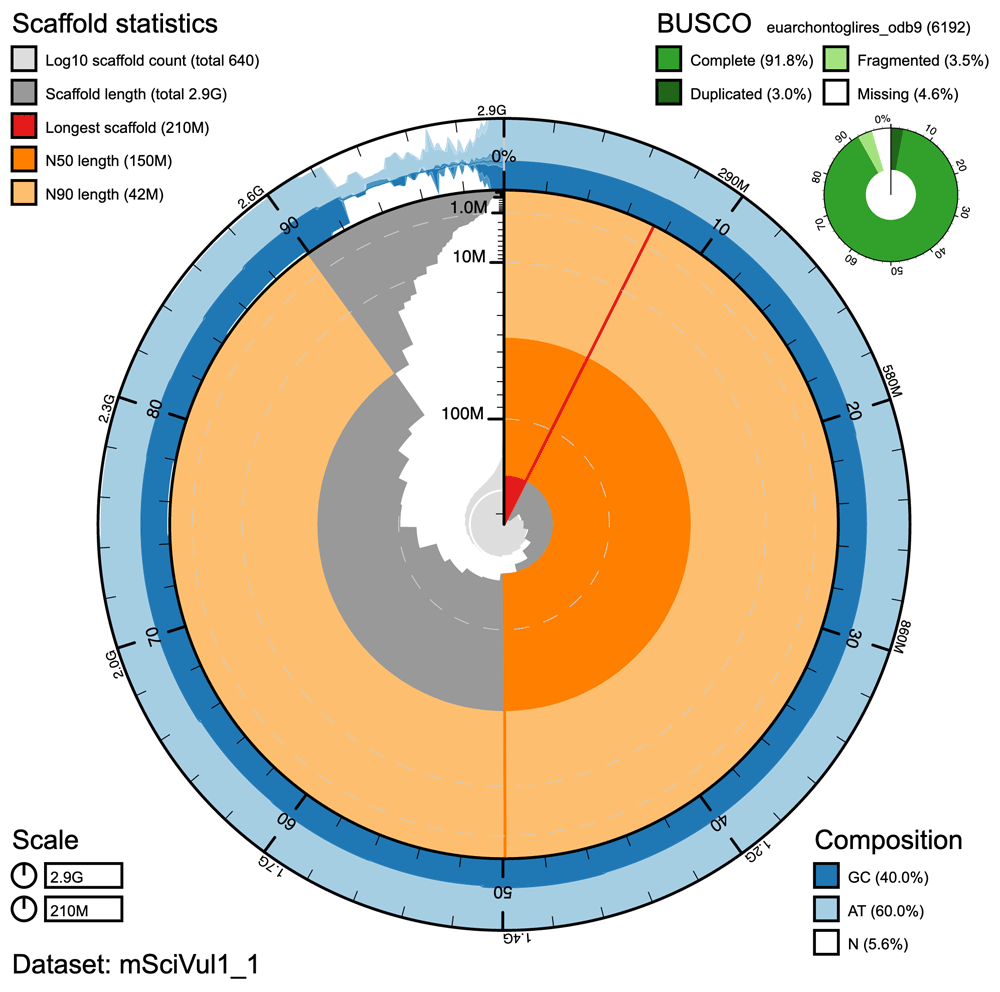
We announce the completion of each Tree of Life genome assembly with a Wellcome Open Research micropublication we call a Genome Note. Genome Notes summarise the origin of the specimen that was used for sequencing, the methods used to extract and sequence the genetic material and the bioinformatic processes that were used to assemble and fine-tune the genome sequence to high quality. They provide assessment of the quality of the genome sequence, using descriptive statistics and figures that demonstrate its quality and accuracy. However, they do not provide any formal analysis of the genome.

In addition to announcing to the world the compilation and availability of the genome, these notes provide citable credit for all involved in the generation of these sequences, from field collectors to infrastructure coordinators, ensuring that the vital contributions of everyone involved in the projects are recognised. These types of brief publication are now common for reporting the genomes of bacteria, viruses and organelles (such as, for example, the American Society for Microbiology’s “Microbiological Resource Announcements”), but the sequences of more complex species have usually been published as more in-depth, analytic research articles.
The sheer number of genomes that are being generated by Tree of Life means that writing a traditional research paper for each species is implausible and would only serve to delay the dissemination of these data. We at Tree of Life want to make these sequences public immediately so that they can be used by researchers at once, rather than embargoing them for years, restricting their utility while we complete analyses and then wait for the editorial boards of traditional journals to decide whether (and then when) they want to publish our work.
Publishing Genome Notes with Wellcome Open Research ensures that the data are available for reuse by all immediately without restriction, maximising their impact. The Wellcome Sanger Institute and Wellcome Open Research have iron-clad Open Data policies and stipulate adherence to the FAIR Data Principles. These policies make Wellcome Open Research a natural fit for us at Tree of Life, where we seek to publish all data, underlying and incidental, immediately and entirely openly. For example, we openly expose quality assessments and other metrics for our raw data and interim progress though ToLQC, and Darwin Tree of Life Data Portal tracks species as they progress through the Tree of Life pipeline and links to all of the data as they become available in the European Nucleotide Archive and in the Ensembl genome database. The rapid publication model of Wellcome Open Research allows the genome sequences to be formally published and shared as quickly as possible, ensuring that researchers are aware they can make use of these data and replicate the methods used in their generation almost instantly.
One of the interesting aspects of this project with Wellcome Open Research is to increase the efficiency in which these genome notes can be prepared and published. F1000, who powers the Wellcome Open Research publishing platform, are developing a new technology to provide a quicker automated route to publication. This means that Genome Notes will ultimately be formulated ‘straight from the sequencer’ and be supported with a combination of machine and community review. More on this very soon!
We hope that the Tree of Life Gateway will serve one additional purpose. As well as announcing the availability of practices and data associated with the programme and providing citable credit for all participants, we also want this Gateway to demonstrate to others that immediate and open data sharing is not only possible but essential if projects of this scale are to have the greatest possible impact.
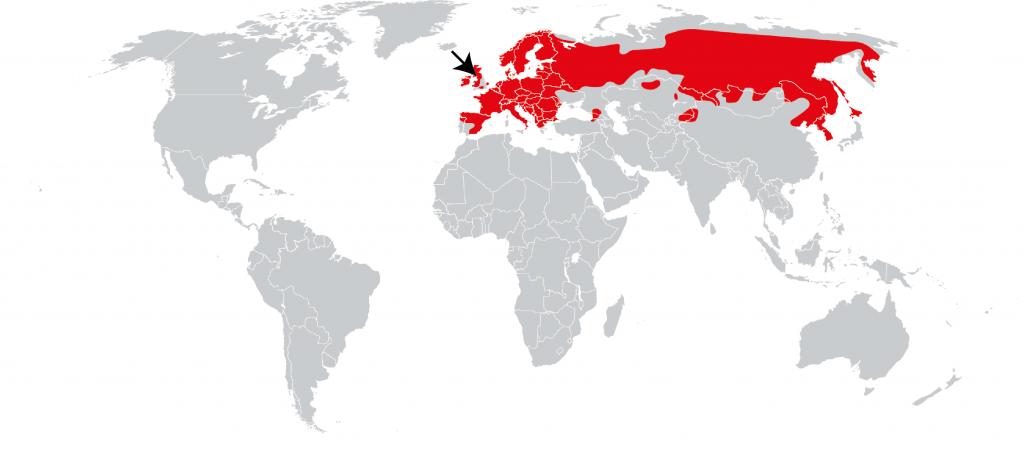
The species we are sequencing in the Darwin Tree of Life and other projects are not “ours” to own, but part of a global patrimony. They have wide distributions, and are often found in many countries or bioregions. By sequencing an individual for a species from a single location (for example the Lancastrian red squirrel presented in one of our first Genome Notes) we are providing a reference for the whole of that species, across its range (the red squirrel is found from Ireland and Portugal to the Kamchatka Peninsula of Russia, Korea and Japan).
We believe that it is through openness and transparency that the worldwide community of researchers and the lay public can be best engaged with projects such as this, and it is only through engagement that we can maximise the impact of the Tree of Life programme and its output.
The gateway launches with seven published Genome Notes. Among these are the majestic Golden Eagle (Aquila chrysaetos), measures are being taken to secure populations and help increase their numbers, but the species remains to be threatened by illegal shooting and poisoning. Also, the native red squirrel (Sciurus vulgaris), a species that has been largely displaced by the American grey squirrel and declining because of the fatal Squirrelpox virus that grey squirrels carry. Make sure to browse through the other genomes, including the bat, otter and rat.