Plant genomics puzzles: many mitochondrial genomes
Plants are some of the most genomically baffling organisms around. As a result, when Darwin Tree of Life launched it was always more likely that our first genome assemblies would be of butterflies rather than bluebells!
Nevertheless our scientists have been busy picking apart the peculiarities of plants, and making some exciting breakthroughs in the process.
Below we explore one example: assembling the multiple genomes that exist within the mitochondria of the common oak (Quercus robur) – the first plant to appear as a published Genome Note.
What usually happens in other organisms?

As you may remember from school, mitochondria are small structures (organelles) within cells that are involved in energy production.
While most of the DNA in eukaryotic organisms (animals, plants, fungi and protists) is contained in the cell nucleus, the mitochondria have a little sequence of DNA all to themselves.
In animals, this mitochondrial DNA is typically formed in a simple ring, much like that of a bacteria. This is thought to be the legacy of early eukaryotic cells joining with bacteria around one and a half billion years ago.
For each animal species, there is mostly a single, identical mitochondrial genome sequence across all cells. The ways in which these can change are limited – although it is worth noting that when they do change this can lead to serious mitochondrial diseases.
Why are plants more complicated?
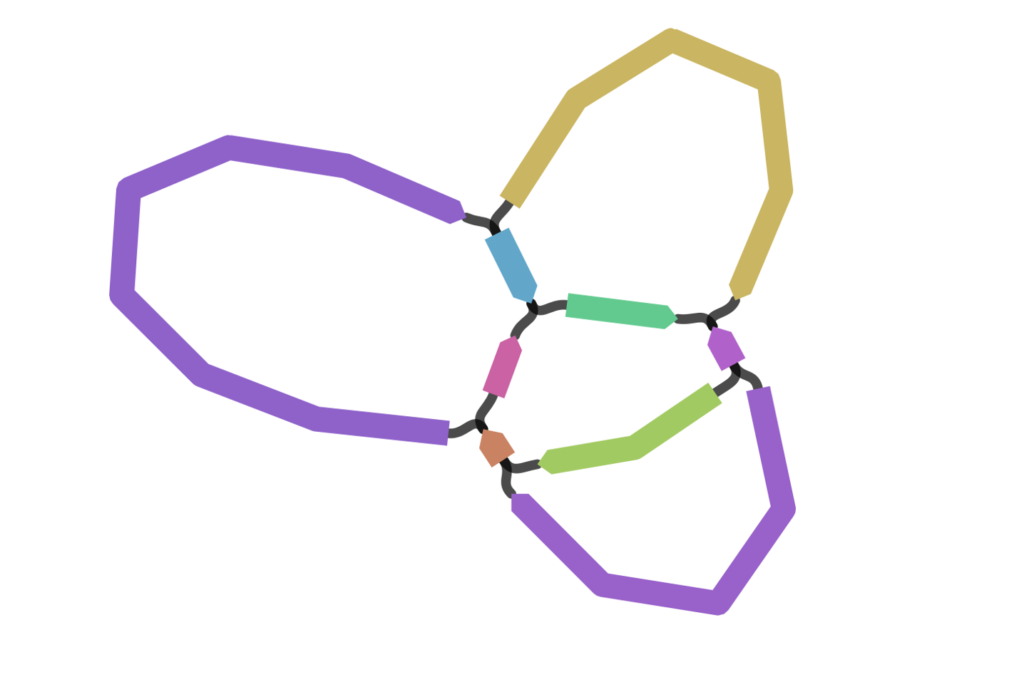
Plants, on the other hand, have several different sequences of DNA in their mitochondria. Scientists think that this happens because different sections of the mitochondrial DNA can bond with each other in a series of different ways in order to make up multiple versions of the full-length sequence.
The region where the bonding takes place is full of lots of repeats of the same small bits of DNA sequence. This gives multiple options for bonding when the cell divides and copies its DNA over.
This means one individual plant can have several completely different sequences for mitochondrial genomes – even within the same tissue from the same part of the plant. In the sample we took from our oak we found three different mitochondrial sequences all occurring in roughly similar amounts.
Was this unexpected?
It is the first time this has been studied in the common oak. But plant genomicists are increasingly noticing this in other species especially due to the development of long-read DNA sequencing technology in recent years.
Short-read sequencing has a hard time picking up these differences. The long-read technology used by Darwin Tree of Life and elsewhere is giving us a much more detailed picture of what’s going on with plant genomes, which previous oak assemblies even a few years ago would not have been able to do.
What does this mean for the future?

Now we have cracked the first of these species with multiple mitochondrial genomes, our scientists are looking at ways to automate and speed up the process. Building the software to do that takes lots of time and effort in itself – but saves much more in a long-term project like ours!
In the meantime, we have also learned plenty of important lessons from the oak genome that we can use to assemble the next plants for the project.
After the sequencing itself, we also need to think about a standard way to present these strangely shaped DNA sequences. We hope that might help reduce further confusion as researchers across the globe study this fascinating phenomenon.
This article was written following an interview with Marcela Uliano-Silva, senior bioinformatician at the Wellcome Sanger Institute.
It is the first in a series on the idiosyncrasies of plant genomes – and the ground-breaking work done by Darwin Tree of Life scientists to assemble them.